
|
Sample |
X 测序平台 |
N测序平台 |
DNBSEQ平台 |
|
Raw bases (Mb) |
99998.92 |
100001.72 |
100236.61 |
|
Clean bases (Mb) |
96314.26 |
98955.15 |
99886.02 |
|
Mapping rate (%) |
99.61 |
98.68 |
99.47 |
|
Unique rate (%) |
87.18 |
86.41 |
93.31 |
|
Duplicate rate (%) |
9.65 |
10.15 |
3.02 |
|
Mismatch rate (%) |
0.8 |
0.51 |
0.48 |
|
Average sequencing depth (X) |
29.08 |
29.52 |
32.8 |
|
Coverage (%) |
99.06 |
99.06 |
99.1 |
|
Coverage at least 4X (%) |
98.57 |
98.43 |
98.62 |
|
Coverage at least 10X (%) |
97.77 |
97.2 |
97.67 |
|
Coverage at least 20X (%) |
91.8 |
89.45 |
92.97 |
已发表文章结果显示,BGISEQ-500自主平台与HiSeq 2500测序平台变异检测的精准度(Precision)和敏感度(Sensitivity)相当[2]。
表2 BGISEQ-500与HiSeq 2500变异精准度和敏感度比较[2]
|
SNP |
BGISEQ-500 |
HiSeq 2500 |
|
Precision |
99.78% |
99.86% |
|
Sensitivity |
96.20% |
96.60% |
DNBSEQ平台变异结果与Illumina Human Omni基因分型芯片评估,结果表明罕见突变检出率高,且检出的罕见突变与芯片分型结果的一致性高。
表3 DNBSEQ平台 30X rare SNP detection rate
|
Genotyping chip |
MAF |
NO. of rare SNP |
NO. of detection |
NO. of concordance |
检出率 |
一致率 |
|
OMNI |
< 2% |
7414 |
7142 |
7132 |
96.33% |
99.86% |
|
OMNI |
< 1% |
3151 |
3025 |
3018 |
96.00% |
99.77% |
|
OMNI |
< 0.5% |
1129 |
1075 |
1070 |
95.22% |
99.53% |
DNBSEQ测序仪利用独特的DNA纳米球(DNB)技术,仅使用单个index就实现了前所未有的0.0001%至0.0004%低样本错误分配率。用水代替DNA,加入index,增加空白对照,DNB测序平台发生错误匹配的概率为36 million reads分之一,即0.0000028%[3]。
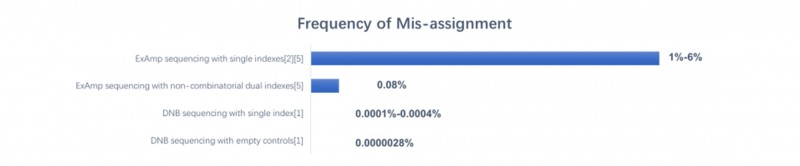
图3 不同测序技术的index hopping比例
DNBSEQ平台WGS数据来源样本种类多样,其中包含福尔马林固定石蜡包埋( Formalin Fixed and Paraffin Embedded,FFPE)样品、单细胞样品、血液样品、基因组DNA样品、唾液样品、常规冷冻保存的新鲜组织样品等。常规基因组建库测序成功率为99%,对于降解样品如FFPE等,建库测序成功率也在90%以上。
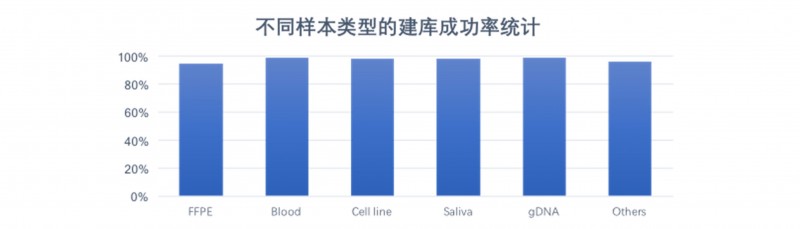
图4 DNBSEQ平台不同类型样本交付成功率
PCR-free建库 + DNB (DNA纳米球)核心测序技术,为您还原真实的全基因组序列。PCR-free WGS 高质量InDel从75%提升到86%,而低质量InDel从12%降低到3%[4],PCR-free建库方法可明显提高InDel calling的精准度和敏感度。
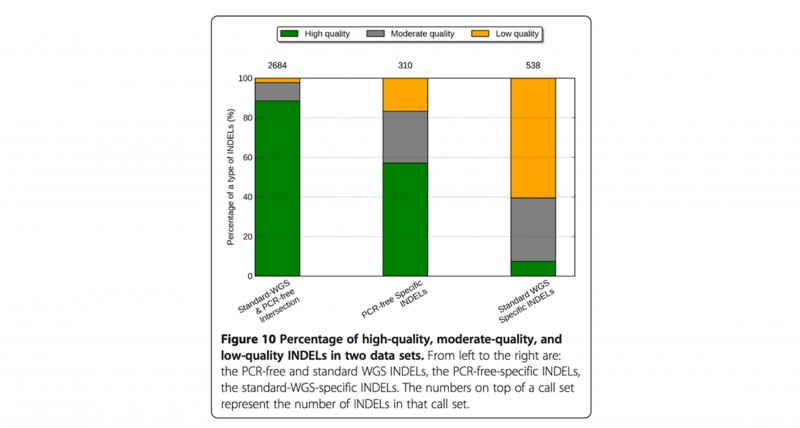
图5 高质量、中等质量和低质量InDel在不同建库方法的分布
参考文献
[1] Drmanac R, Sparks A B, Callow M J, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays.[J]. Science, 2010, 327(5961):78-81.
[2] Jie Huang, Xinming Liang, Yuankai Xuan, et al. A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience, 2017.
[3] Li Q, Zhao X, Zhang W, et al. Reliable Multiplex Sequencing with Rare Index Mis-Assignment on DNB-Based NGS Platform. bioRxiv, 2018: 343137
[4] Han F, Wu Y, Narzisi G, et al. Reducing INDEL calling errors in whole genome and exome sequencing data[J]. Genome Medicine,6,10(2014-10-28), 2014, 6(10):89.